 |
| Courtesy: ACS Nano DOI: 10.1021/acsnano.7b03073 |
For any couple who has witnessed an amniocentesis with WIDE eyes (as I did), this advance should be a welcome relief.
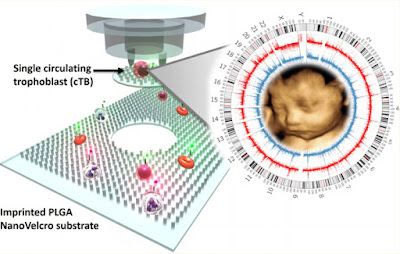
Circulating fetal nucleated cells (CFNCs) in the blood of pregnant women is an ideal source of fetal genomic DNA that can be used for prenatal diagnostics. However, the problem is that there are only a very small number of CFNCs in maternal blood. A team of researchers in the US, China and Taiwan has now developed nanoVelcro microchips that can effectively enrich a subcategory of CFNCs, namely circulating trophoblasts (cTBs) in blood samples. These cTBs can then be isolated using a laser microdissection technique for subsequent genetic testing.
Current prenatal tests for diagnosing foetal genetic abnormalities rely on invasive, “harvesting” procedures, such as amniocentesis and chorionic villus sampling. Although highly valuable, they can increase the risk of miscarriage. Whole foetal cells circulating in an expectant mother’s blood could also provide important information on foetal DNA since they contain entire genomes, but until now it has been very challenging to capture these cells because they are only present in small quantities.
The new nanoVelcro microchips developed by Hsian-Rong Tsung of the California NanoSystems Institute at the University of California at Los Angeles and colleagues can effectively enrich cTBs from blood samples. These cells can then be isolated using a technique called laser capture microdissection (LCM) for subsequent genetic testing.
The researchers (who initially developed their microchips for detecting low concentrations of tumour cells circulating in blood) made their devices by nano-imprinting them on a spin-coated PLGA substrate (see image). To enrich the cTBs, they grafted a biotinylated anti-EpCAM (which is a trophoblast surface marker) onto the imprinted nanoVelcro.
For the genetic characterization, they isolated at least three individual cTBs and pooled these together in a 0.5 mL polymerase chain reaction (PCR) tube for whole genome amplification (WGA). They then subjected the resulting amplified DNA to so-called array comparative genomic hybridization (array CGH) and short tandem repeat (STR) assays.
NanoVelcro microchips for prenatal testing, Belle Dumé, Nanotechweb.org
Comments